
The BELKA dataset
This is the dataset provided for the BELKA competition, which Leash Biosciences hosted on Kaggle in the summer of 2024. It is roughly 100M small molecules (down from 133M after splitting) from DNA-encoded chemical libraries screened against 3 protein targets (BRD4, EPHX2/sEH, and ALB/HSA). More detailed descriptions of the targets, including links to target crystal structures for structure-based predictions, can be found here.
DELs
The vast majority of chemical matter in this dataset is a triazine-based 3-cycle library designed to superficially resemble DEL-A in (1) and this library is called AMA014. AMA014 was designed and manufactured by AlphaMa.
The orthogonal DEL in the library split is a non-triazine 3-cycle library designed by Leash to resemble kinase inhibitor chemistry and is called kinase0, or kin0 for short.
ML-Ready: To benchmark against the BELKA competition
For the contest, we provided binary bind-or-no-bind labels for each small molecule pair. We also provided data splits: 1) a random molecule split, 2) a split where a central core (a triazine) is preserved but there are no shared building blocks between train and test, and 3) a library split. The library split is a series of screens with the same targets against an entirely different DEL with different building blocks, different cores, and different attachment chemistries. The library split was included in the test set only, to test true out-of-distribution predictions.
The dataset that you're looking at now is identical to the dataset in the Kaggle competition in order to enable those who want to benchmark methods against it.
Raw Readouts: To try new methods of hit-calling and more
Maybe you want to know how replicates help, or if multiple rounds of selection is a good idea with AMA014, or what background conditions might be best. This dataset is for you!
To allow exploration of additional hit-calling methods and the contributions of various aspects of experimental design to enriching those hits, we have has also provided the raw/normalized sequencing counts and experimental details of each replicate. These can be found in a public cloud bucket at https://storage.polarishub.io/belka-v1-raw/.
Please note that the full raw dataset uses ~600GB in its compressed form!
Details about the raw readouts
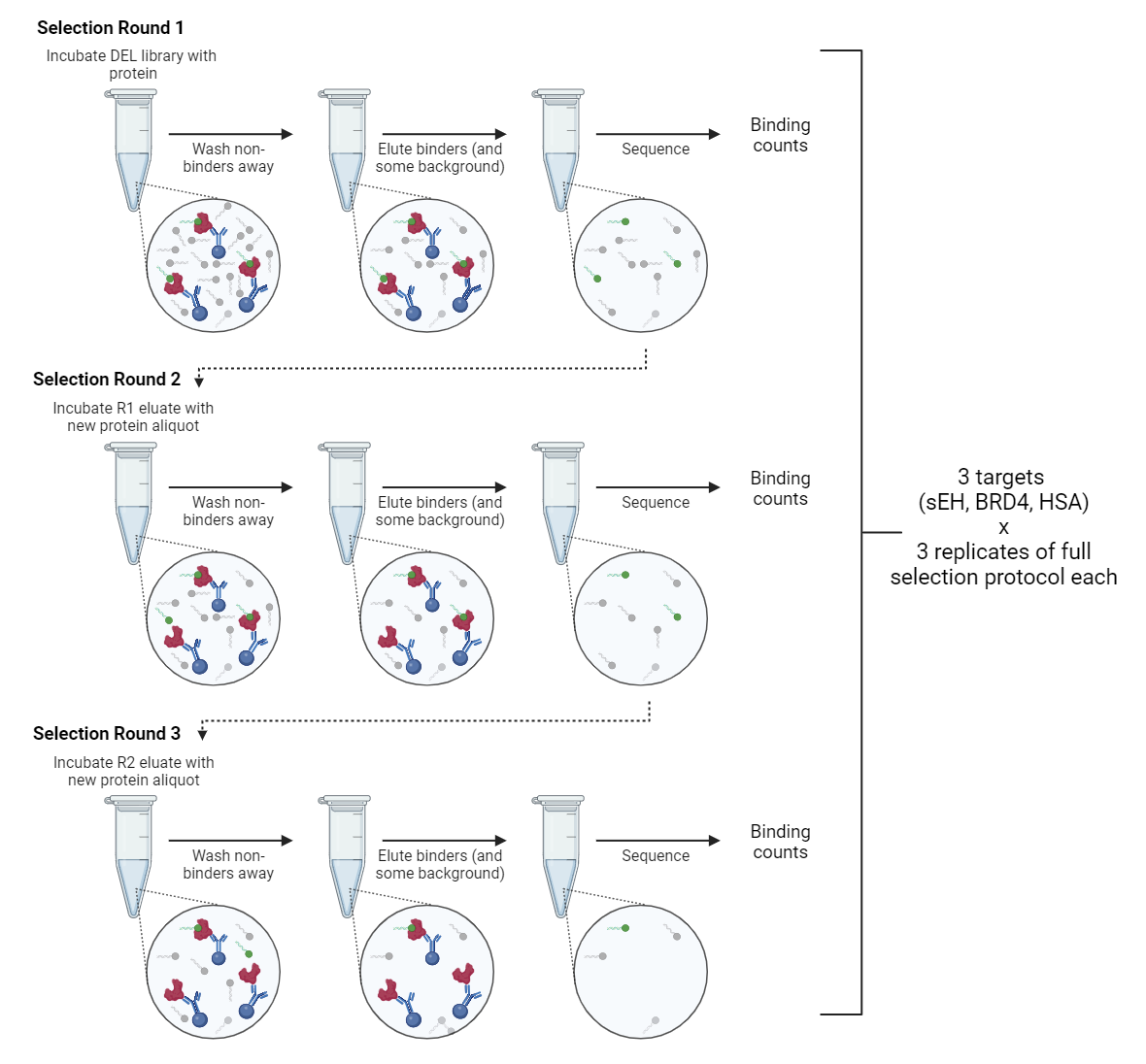
Rounds of selection:
A common DEL screening practice is to combine the library with the protein target, isolate the DEL/target complexes, elute the bound DEL (here, we did it with heat), and repeat the selection with fresh target protein (Figure 1). The reasoning is that extra rounds select for only the tightest binders, making the most efficient use of sequencing capacity (1). To test this method, we performed 3 rounds of selection on the triazine library per target protein and sequenced a small portion of each round. Kinase0 is a far smaller library, and only a single round of selection was performed on it.
Number of replicates
Each series of selection rounds was performed in triplicate for AMA014. Kinase0 screens were performed in duplicate with a single round of selection.
Columns in the individual experiment dataset
molecule_id: unique identifier per moleculesmiles: SMILES representation of the full moleculebb1_smiles, bb2_smiles, bb3_smiles: SMILES representation of the individual building blocks that, when combined using SMARTS reactions, produce the full molecule (also with a triazine core, in the case of AMA014)bcj_id: binding count job, an id created internally at Leash to track computational counting jobsraw_counts: read counts obtained from the sequencing datacounts_per_billion: read counts normalized by the size of the entire sequencing run and scaled to one billion reads
Conventions
The raw data can be found in folders following the sample label naming convention provided below, one folder per selection round/replicate.
Conventions for Target Screens:
Sample labels for AMA014:
[target]-[replicate]-[selection round]
For example:
- HSA-A-R1/ is HSA, replicate 1, first selection round (see
Rounds of selection below)
- SEH-C-R2/ is sEH, replicate 3, second selection round
Sample labels for kinase0:
[target]-[kin0]-[replicate]
For example:
- SEH-Kin0-S1/ is sEH, kinase0, replicate 1, first (and only) selection round
Conventions for negative controls:
Negative control conditions.
One approach to identifying hits is by comparing the relative enrichment of sequenced barcodes over a background distribution. The distribution of the DEL by itself is such a background; an alternate background would be to mix the DEL and the magnetic beads used in selection to identify a bead-only distribution. We are providing both.
Sample labels for DEL-only negative controls:
We sequenced the DELs alone in duplicate to understand the background distribution of the libraries.
DEL-A, DEL-B are replicates of AMA014 alone.
DEL-Kin0-S1, DEL-Kin0-S2 are replicates of kinase0 alone.
Sample labels for bead-only negative controls:
To compare bead-only signal against the AMA014 selections, we used His Dynabeads from Thermo in triplicate. The Dynabeads were used in the protein-target selections of AMA014. His-A, His-B, and His-C are replicates of Dynabead-only selections, one selection round only.
To compare bead-only signal against the kinase0 selections, we used EDTA-compatible Magbeads from Thermo in duplicate. The Magbeads were used in the protein-target selections of kinase0. Some building blocks in kinase0 were strong Dynabead binders, which is why the Magbeads were used in these screens.
HisEDTA-Kin0-S1 and HisEDTA-Kin0-S2 are replicates of Magbead-only selections, one selection round only.