Tuesday, December 3, 2024
Antiviral Competition: Advancing Open Science with ASAP Discovery and OpenADMET

Small molecules continue to be the backbone of drug discovery, accounting for ~75% of drugs approved by the FDA over the last decade. Their versatility, oral bioavailability, cost-effectiveness, and proven scalability in manufacturing make them an enduring cornerstone of the pharmaceutical industry.
ASAP Discovery is an NIH-funded consortium leveraging open science for antiviral drug discovery, aiming for equitable and affordable global access to effective antivirals. ASAP has pursued several programs and targets, with the most advanced being ASAP’s dual SARS-CoV-2 and MERS-CoV main protease (Mpro) program, reaching preclinical candidate nomination. You can see a full list of ASAP’s programs on the website.
ASAP Discovery is passionate about open science and has put a huge amount of effort into sharing its outputs in a digestible way with the community. For example, if you navigate to ASAP’s website, the drug discovery pipeline is fully interactive for users. Clicking any filled box will navigate you to the continuously published data for those experiments, and experimental protocols used.

The Challenge
Blind challenges are a critical tool to evaluate and advance the current state of the art in computational molecular sciences, see here for some recent blind challenges in drug discovery. Sharing of these sorts of learnings has led to innovations such as AlphaFold2 as with the CASP sequence of folding challenges.
As part of its open science mission, the ASAP Discovery Consortium is conducting a computational methods challenge encompassing several modalities critical to small molecule drug discovery. This challenge will be run in collaboration with OpenADMET, which is a new ARPA-H funded project under the Open Molecular Software Foundation (OMSF).
ASAP Discovery is approaching a patent disclosure for its preclinical candidates for these two Mpro programs. There is a batch of data in these projects that ASAP Discovery has not publicly disclosed; this will be the blind test data of this challenge. The challenge will mirror some of the real-world drug discovery challenges that ASAP has had to overcome in the last three years. You will be working with active and real drug discovery data normally restricted to large pharmaceutical companies!
Competition Details
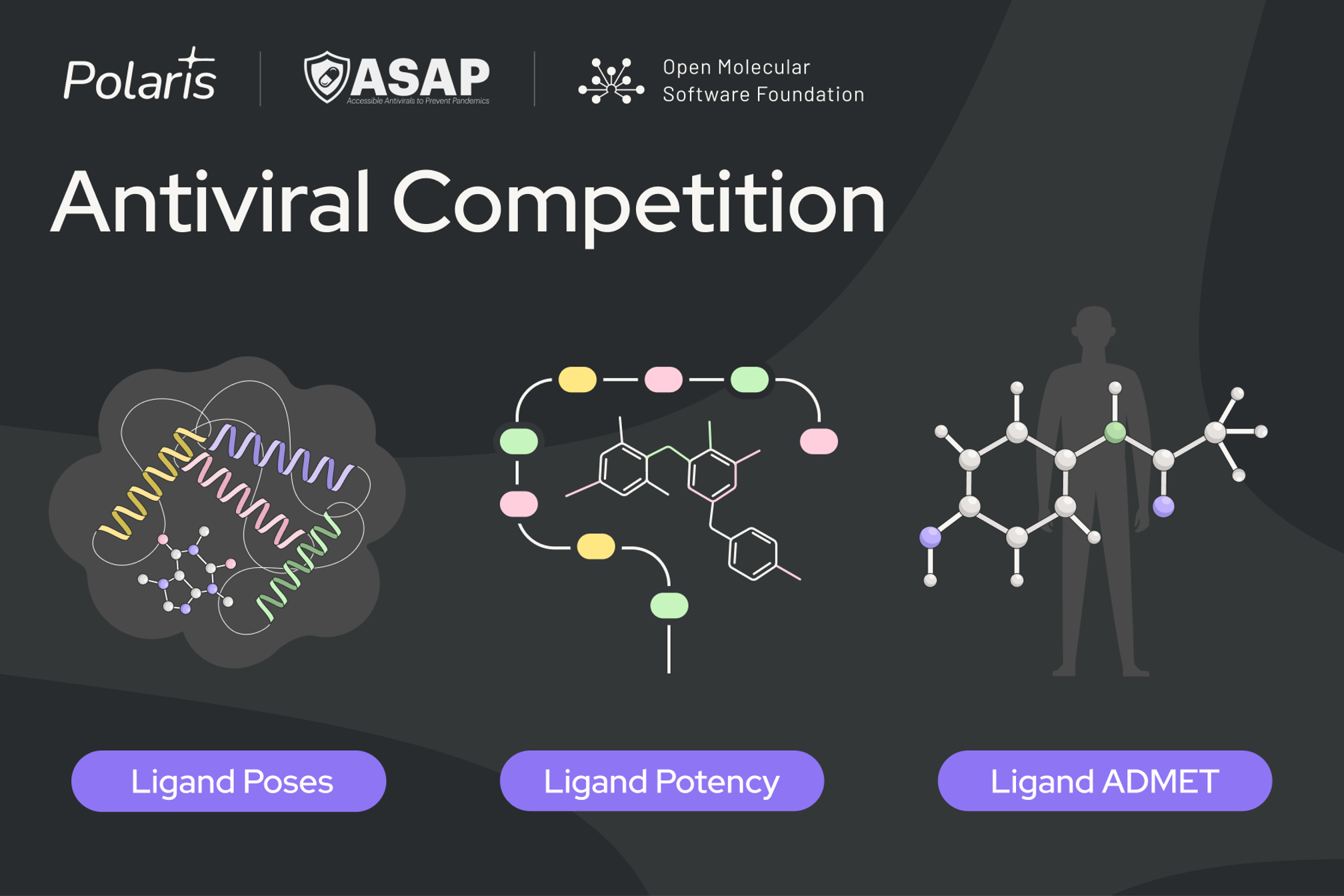
This challenge will be composed of three sub-challenges:
- 🧩 Ligand Poses: ASAP has produced a large volume of X-ray crystallography data over its years of operation. Along this trajectory, SARS-CoV-2 Mpro was structurally enabled much earlier than MERS-CoV. This sub-challenge will recreate that situation. Given a training set of SARS-CoV-2 Mpro X-ray structures, participants will be asked to predict poses of a test set of compounds for MERS-CoV Mpro. The crystallography experiments for this sub-challenge were performed by the University of Oxford and Diamond Light Source. See here for the crystallography conditions.
Access the sample data today. - 💥 Potency: Given a training set of dose-response fluorescence potency data for both targets (SARS and MERS Mpro), participants will be challenged to predict potencies for a blind set of compounds for both targets. The assays for this sub-challenge were performed by the Weizmann Institute of Science. See here for the experimental conditions.
Access the sample data today.
- 🧪 ADMET: This sub-challenge will consist of multiple ADMET endpoints. Participants will receive training data for all endpoints and will be asked to predict the same endpoints for a blind set of compounds. The assays for this sub-challenge were performed by Bienta.
Access the sample data today.
Judging Criteria
We welcome submissions of any kind, including machine learning and physics-based approaches. You can employ pre-training approaches as you see fit. You are also free to reuse data from one portion of the challenge for others if it will assist you (e.g ligand poses can be used to assist binding affinity predictions). In the open science spirit of ASAP Discovery we would love to see open code showing how you created your submission if possible. You can enter as many times as desired, and the challenge will be judged based on the judging criteria outlined here.
- Potency: we will evaluate your submission using the mean absolute error of the predicted endpoint.
- ADMET endpoints: we will evaluate your submission using MAE on the log transformed endpoints, after clipping to the strictly positive detection limit. This minimises the effect of massive outliers on the MAE. For LogD which is already in log units we will just use MAE directly.
- Ligand posing: we will evaluate your submission using % of poses with a RMSD<2A compared to the crystallographic pose.
Prizes
We’ll be offering Polaris merch packs to the top three performing teams for each sub-challenge. We will also be writing our conclusions up as a paper, to which all submitting teams are invited to share co-authorship.
Post-challenge virtual workshop
Participants with considerable performance or learnings will have the opportunity to present their work at a special blind-challenge workshop to share learnings, hosted by the NIH ASAP Open Science Forum.
A special issue in the Journal of Chemical Information and Modeling
We encourage everyone participating in this challenge to share a preprint - we will track these during the challenge so that participants can compare learnings on-the-fly - that describes their approach, results, performance and learnings from comparing their approach with other groups. We are working with the editors of Journal of Chemical Information and Modeling (JCIM; a high-impact journal in chemical informatics and molecular modeling) on a collection of papers/ special issue that will report on the breadth of learnings and outcomes from this challenge.
Timelines

How to get started
The best way to get started is to browse some of the data available on Polaris already! Have a play around with the sample data (structures, potency, ADMET) and get a feel for what the challenge data will look like. Try building some models or making some predictions and see what works. We also encourage people to form teams with your colleagues, company, workplace, and pets! If you would like to meet other possible team members, come and post on our Discord channel! Once the challenge officially starts on Jan 13, we hope to see you all on the leaderboard!
Next Steps
If you would like more information or would like to see current discussions on the ASAP Discovery competition, come and hang out on the Polaris Discord channel! We will also be hosting regular office hours starting in Jan 2025, with details to follow on Discord. We are very very excited to see what the community can come up with! Blind challenges have the potential to transform modeling and machine learning for drug discovery, come be a part of it!
You can also stay updated on the latest developments with the competition by signing up to the Polaris mailing list!