🌹 Runs N' Poses 🌹
A protein-ligand co-folding prediction dataset and benchmark
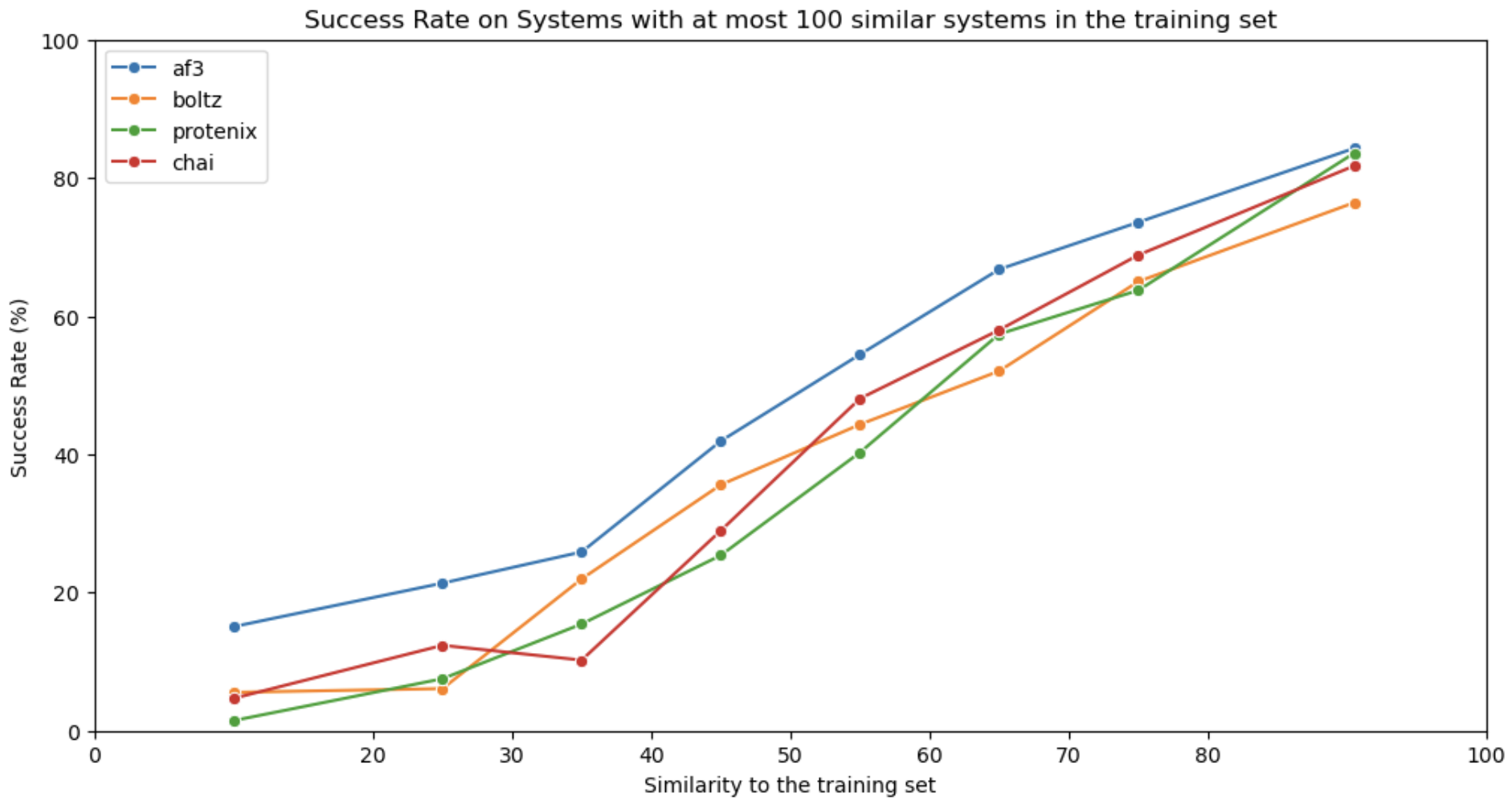 Figure last updated on February 4th, 2025
Figure last updated on February 4th, 2025
A protein-ligand co-folding benchmark with 2,073 high-resolution protein-ligand systems released after 30 September 2021 with at most 100 similar ligands in the train set. The test set is further split in multiple subsets with increasing distance to the train set to test for generalization. Performance is measured using a compound metric of BiSyRMSD and LDDT-PLI.
See the GitHub repository for more details on how these methods were run and evaluated.
Metrics
We measure the following 4 performance measures
success_rate: The percentage of predictions with a LDDT-PLI > 0.8 and a BiSyRMSD < 2.rmsd_success_rate : The percentage of predictions with a BiSyRMSD < 2.mean_lddt_pli : The average LDDT-PLI.coverage : The percentage of systems that the method could produce a prediction for.
Split
Train
We don't provide the train set here. The train set should only include PLI systems from before 30 September 2021.
Test
The test set consists of 2,073 high-resolution protein-ligand systems released after 30 September 2021 with at most 100 systems with similar ligands in the train set (same as Figure 1B in the paper cited below). It is further divided in 8 subsets, binned by their similarity (as defined by sucos_shape_pocket_qcov) to the training set:
- Test 0 to 20 (
n=76)
- Test 20 to 30 (
n=106)
- Test 30 to 40 (
n=149)
- Test 40 to 50 (
n=272)
- Test 50 to 60 (
n=301)
- Test 60 to 70 (
n=364)
- Test 70 to 80 (
n=328)
- Test 80 to 100 (
n=477)
Citation
If you use this dataset in your research, please cite Runs N' Poses and PLINDER:
Škrinjar, P., Eberhardt, J., Durairaj, J., & Schwede, T.
"Have protein-ligand co-folding methods moved beyond memorisation?"
bioRxiv (2025): 2025-02
Durairaj, Janani, et al.
"PLINDER: The protein-ligand interactions dataset and evaluation resource."
bioRxiv (2024): 2024-07.