Background
ADME@NCATS is a resource developed by NCATS to host in silico prediction models for various ADME (Absorption, Distribution, Metabolism and Excretion) properties. The resource serves as an important tool for the drug discovery community with potential uses in compound optimization and prioritization. The models were retrospectively validated on a subset of marketed drugs which resulted in very good accuracies.
Data that were used for developing the models are made publicly accessible by depositing them into PubChem database. In some instances, when complete data cannot be made public, a subset of the data are deposited into PubChem. Links to the PubChem assays can be found in the individual model pages. The users are highly encouraged to use these data for development and validation of QSAR models.
Assay Information
Hepatic metabolic stability is a key pharmacokinetic parameter in drug discovery. Metabolic stability is usually assessed in microsomal fractions and only the best compounds progress in the drug discovery process. A high-throughput single time point substrate depletion method in rat liver microsomes (RLM) is employed at the National Center for Advancing Translational Sciences (NCATS) as a Tier 1 assay. Between 2012 and 2020, RLM stability (in vitro half-life) data was generated for ~24,000 compounds from more than 250 NCATS projects that cover a wide range of pharmacological targets and cellular pathways. Data for ~2500 compounds along with the global prediction models are publicly available.
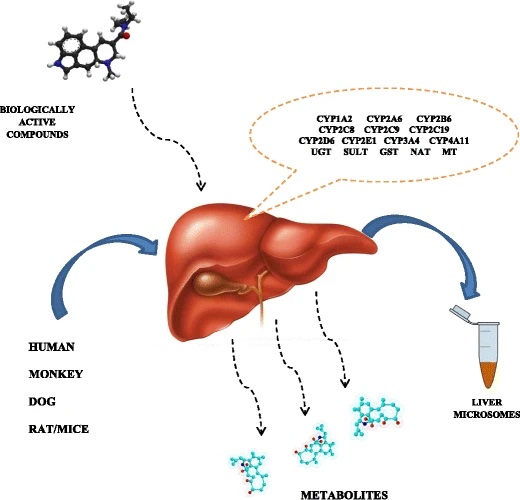
Image is from this paper. Biologically active compounds can be transformed or destroyed by the action of enzymes in the liver. Microsomes are small membrane bubbles (vesicles) that come from a fragmented cell membrane, and can be used as a proxy for how well a drug survives a trip through the liver.
Description of readout:
- PUBCHEM_ACTIVITY_OUTCOME: Corresponds to the phenotype observed. For all compounds with phenotype "stable", PUBCHEM_ACTIVITY_OUTCOME is "active" (class = 1). For all compounds with phenotype "unstable", PUBCHEM_ACTIVITY_OUTCOME is "inactive" (class = 0).
- PHENOTYPE: Based on the half-life observed. Compound is "stable" if half-life (t1/2) is more than 30 minutes (class = 1); "unstable" if t1/2 is less than 30 minutes (class = 0).
- HALF-LIFE: Rat liver microsome stability (t1/2), in minutes. >30 minutes has been converted to 31.
Optimization objective: A drug with high hepatic metabolic stability is often desirable as it can result in a more sustained therapeutic effect with lower dosing frequencies. However, excessive metabolic stability can lead to drug accumulation and potential toxicity. There may not be a universally applicable optimal range for hepatic metabolic stability, the goal is to achieve a balance that maximizes therapeutic benefit while minimizing adverse effects and maintaining an acceptable safety profile.
In the context of this dataset, stable compounds are preferable.
Data resource
Reference: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7693334/
Raw data: https://pubchem.ncbi.nlm.nih.gov/bioassay/1508591